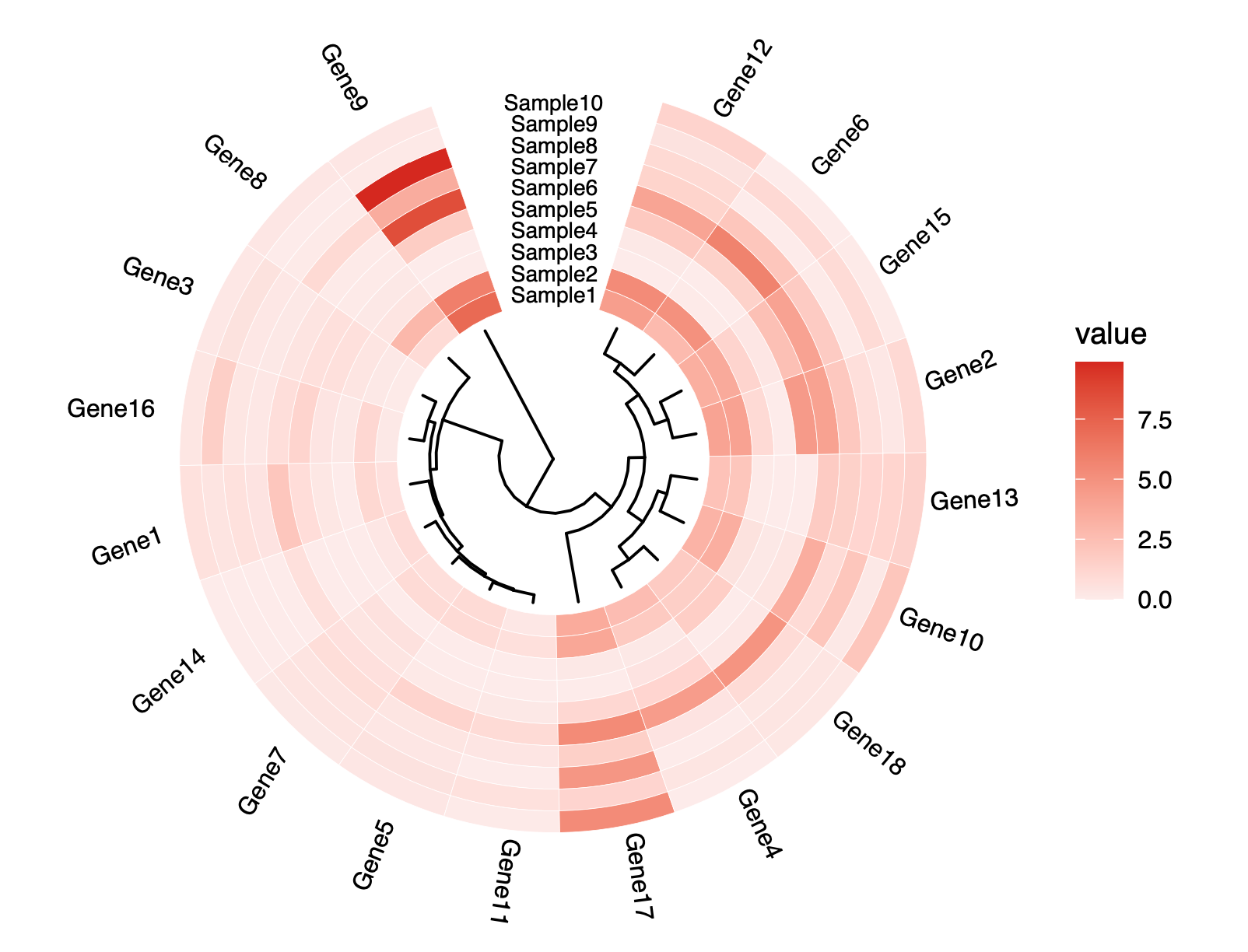
差异基因表法分析

差异基因表法分析
泡泡DESeq2
1 | # |
Deseq2的输出中,Padj实际上是FDR! 一般我们认为:PAdj进行多重检验所使用的方法是hochberg, FDR进行多重检验的方法是Benjamini & Hochberg。所以在代码中我们进行了转换操作。
数据结果说明
name - the feature identity. It must be unique within the column. It may be a gene name, transcript name, or exon - whatever the feature we chose to quantify.
baseMean - the average normalized expression level across samples. It measures how much total signal is present across both conditions.
baseMeanA - the average normalized expression level across the first condition. It measures how much total signal is there for condition A.
baseMeanB - the average normalized expression level across the first condition. It measures how much total signal is there for condition B.
foldChange - the ratio of baseMeanB/baseMeanA. Very important to always be aware that in the fold change means B/A (second condition/first condition)
log2FoldChange - the second logarithm of foldChange. Log 2 transformations are convenient as they transform the changes onto a uniform scale. A four-fold increase after transformation is 2. A four-fold decrease (1/4) after log 2 transform is -2. This property makes it much easier to compare the magnitude of up/down changes.
PValue - the uncorrected p-value of the likelihood of observing the effect of the size foldChange (or larger) by chance alone. This p-value is not corrected for multiple comparisons.
PAdj - the multiple comparisons corrected PValue (via the Hochberg method). This probability of having at least one false positive when accounting for all comparisons made. This value is usually overly conservative in genomics.
FDR - the False Discovery Rate - this column represents the fraction of false discoveries for all the rows above the row where the value is listed. For example, if in row number 300 the FDR is 0.05, it means that if you were cut the table at this row and accept all genes at and above it as differentially expressed then, 300 * 0.05 = 15 genes out of the 300 are likely to be false positives. The values in this column are also called q-values.
falsePos - this column is derived directly from FDR and represents the number of false positives in the rows above. It is computed as RowIndex * FDR and is there to provide a direct interpretation of FDR.
The following columns represent the normalized matrix of the original count data in this case, 3 and 3 conditions.
edgeR
1 | # |
数据结果说明
name - the feature identity. It must be unique within the column. It may be a gene name, transcript name, or exon - whatever the feature we chose to quantify.
baseMean - the average normalized expression level across samples. It measures how much total signal is present across both conditions.
baseMeanA - the average normalized expression level across the first condition. It measures how much total signal is there for condition A.
baseMeanB - the average normalized expression level across the first condition. It measures how much total signal is there for condition B.
foldChange - the ratio of baseMeanB/baseMeanA. Very important to always be aware that in the fold change means B/A (second condition/first condition)
log2FoldChange - the second logarithm of foldChange. Log 2 transformations are convenient as they transform the changes onto a uniform scale. A four-fold increase after transformation is 2. A four-fold decrease (1/4) after log 2 transform is -2. This property makes it much easier to compare the magnitude of up/down changes.
PValue - the uncorrected p-value of the likelihood of observing the effect of the size foldChange (or larger) by chance alone. This p-value is not corrected for multiple comparisons.
PAdj - the multiple comparisons corrected PValue (via the Hochberg method). This probability of having at least one false positive when accounting for all comparisons made. This value is usually overly conservative in genomics.
FDR - the False Discovery Rate - this column represents the fraction of false discoveries for all the rows above the row where the value is listed. For example, if in row number 300 the FDR is 0.05, it means that if you were cut the table at this row and accept all genes at and above it as differentially expressed then, 300 * 0.05 = 15 genes out of the 300 are likely to be false positives. The values in this column are also called q-values.
falsePos - this column is derived directly from FDR and represents the number of false positives in the rows above. It is computed as RowIndex * FDR and is there to provide a direct interpretation of FDR.
The following columns represent the normalized matrix of the original count data in this case, 3 and 3 conditions.